Get The Memo.
Alan D. Thompson
July 2024, updated Dec/2024
Summary
| Organization | Argonne National Laboratory (a US Department of Energy lab near Chicago, Illinois) |
| Model name | AuroraGPT |
| Internal/project name | A derivative model will be called ‘ScienceGPT’ |
| Model type | Multimodal (text, specialized scientific outputs like temp, LiDAR ranges, etc) |
| Parameter count | 7B ➜ 70B ➜ … ➜ 2T |
| Dataset size (tokens) | 10T general data (~40TB) >20T scientific data (~80TB) = 30T tokens total (~160TB) |
| Training time (total) | 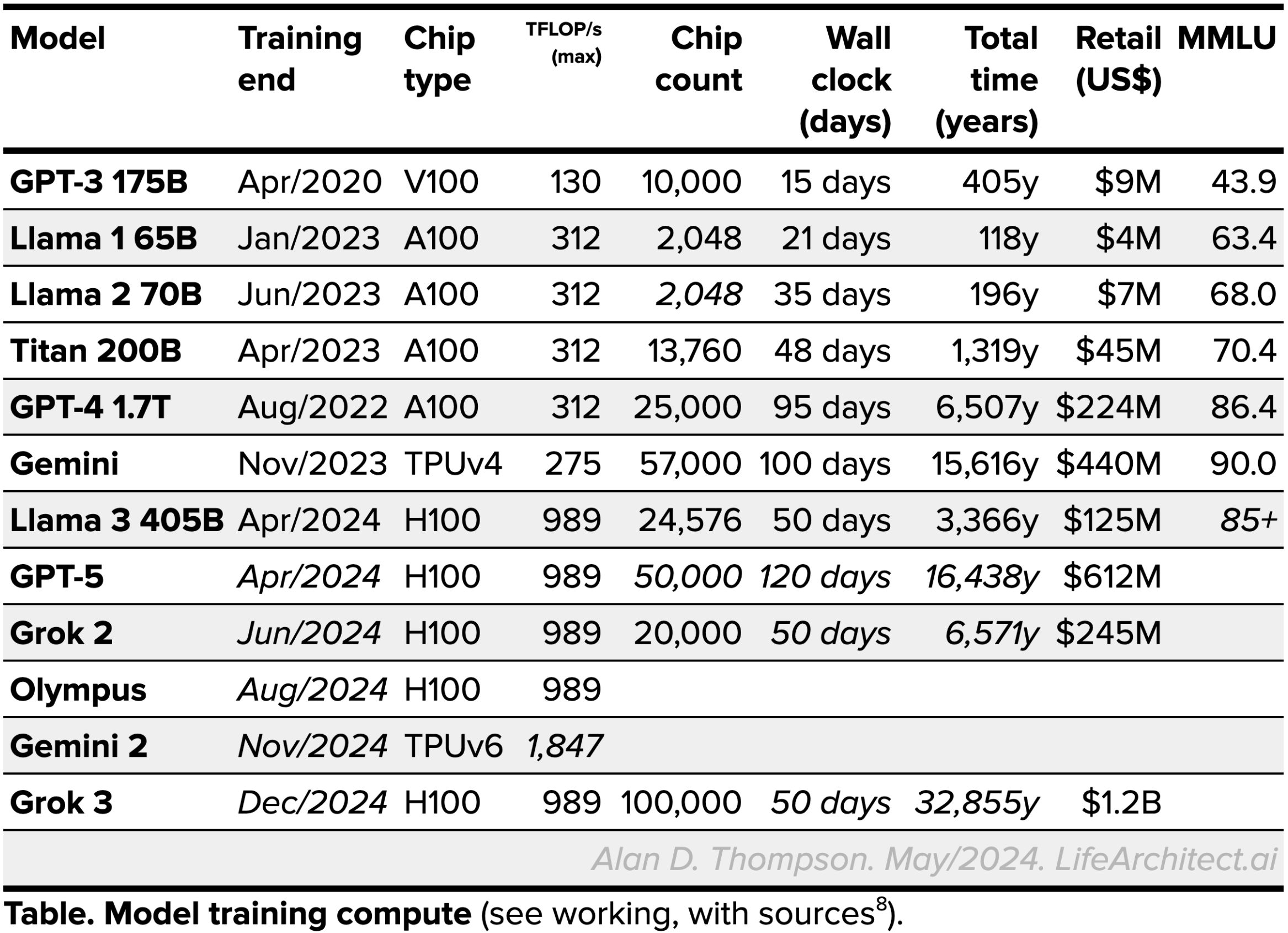 See working, with sources.
See working, with sources. |
| Release date (public) | Alan expects: 2025 |
| Paper | – |
| Playground | – |
AuroraGPT Updates
Jan/2026: New paper published.
 Genesis Mission:
Genesis Mission:
A Comprehensive Analysis of US Science Datasets
Alan D. Thompson
LifeArchitect.ai
January 2026
75+ pages incl extended matter.
9/Sep/2024: Argonne released a slide deck from Euro-Par 2024, showing a lot of information presented in a decidedly scientific format (PDF, 68 pages, 9.6MB, archive via The Memo 14/Nov/2024).
18/Jul/2024: Three initial models to be trained, all small 7B parameters:
- AuroraGPT-7B-P (Ponte Vecchio GPU testing)
- AuroraGPT-7B-A (Aurora)
- AuroraGPT-7B-A-S (Aurora + Science)
Source: Argonne, 18/Jul/2024
13/Nov/2023: Intel, NVIDIA, Microsoft link:
ANL and Intel are in the early stages of testing the hardware before putting the model into full training mode… AuroraGPT is enabled by Microsoft’s Megatron/DeepSpeed… Intel has worked with Microsoft on fine-tuning the software and hardware, so the training can scale to all nodes. (HPCwire)
23/May/2023: Aurora supercomputer details:
With two exaflops of performance, the Intel-powered Aurora supercomputer is expected to beat the AMD-powered Frontier supercomputer, currently the fastest in the world, and take the lead on the Top 500 list of the fastest supercomputers…
21,248 CPUs and 63,744 Ponte Vecchio GPUs, Aurora will either meet or exceed two exaflops of performance when it comes fully online before the end of the year [2023]. The system also features 10.9 petabytes (PB) of DDR5 memory, 1.36 PB of HBM attached to the CPUs, 8.16 PB of GPU memory, and 230 PB of storage capacity that delivers 31 TB/s of bandwidth… (Tom’s)
Dataset
“The mandate of the data team… To accumulate on the order of 20 plus trillion tokens of high quality scientific text and structured data with strong quality control, deduplication.” (Argonne, 18/Jul/2024)
General text data (filtered to 10T tokens)
| Source | Subset | Tokens (unfiltered) |
|---|---|---|
| FineWeb v1 (Common Crawl) | – | 15T total |
| Dolma (3T total incl 2.59T CC, removed for dedupe) |
The Stack | 430B |
| PeS2o [~40M academic papers] | 57B | |
| Project Gutenberg | 4.8B | |
| Wikipedia + Wikibooks | 3.6B |
Scientific data (multimodal, filtered to >20T tokens)
| Source (by unfiltered size) | Tokens (unfiltered) |
|---|---|
| Hardware/Hybrid Accelerated Cosmology Code (HACC) Simulation |
600T tokens* 1.7PB1‘Building on a high-performance 1.7PB parallel file system (to be increased in the near future) and embedded in Argonne National Laboratory’s 100+ Gbps network fabric… Given the size of the raw outputs (up to several PBytes for our largest simulations) it is very difficult (and expensive) to provide access to Level 1 data. Level 2 data has an added layer of processing, which makes it roughly an order of magnitude smaller in size; examples include downsampled particle snapshots, halo catalogs, and halo merger trees. Most scientific results are based on the analysis of Level 2 data and post-processing thereof to generate Level 3 data. Level 3 data includes maps and object catalogs at different wavelengths. In this first data release, we focus on Level 2 data, namely downsampled particle outputs and halo catalogs.’ |
| Bacterial and Viral Bioinformatics (BV-BRC)… | 148T tokens* 418.3TB2‘Total storage used for user data (TB): 418.3’ |
| Energy Exascale Earth System Model (E3SM) | 12T tokens* 34TB3‘The piControl run with 500 years of data size: Published 34T of data, 43742 files’ |
| PubMed (full, not just abstracts or Central subsets) | 600B tokens* 37M docs4‘PubMed® comprises more than 37 million citations for biomedical literature from MEDLINE, life science journals, and online books. Citations may include links to full text content from PubMed Central and publisher web sites.’ =1.7TB* |
| American Society for Microbiology (ASM) | 283B tokens* 800GB=17M docs* |
| Association for Computing Machinery (ACM): Elsevier, Wiley, Springer, CreateSpace, McGraw-Hill, Prentice-Hall, Addison-Wesley, Apress, Stanford, MIT, CMU, IEEE, NASA Langley… |
66B tokens* 4M docs5‘As of March 2020, the DL contains 786,000 full-text articles, and 3.2 million tracked publication records.’=188GB* |
| arXiv.org e-Print archive | 40B tokens* 2.4M docs6‘arXiv is a free distribution service and an open-access archive for nearly 2.4 million scholarly articles in the fields of physics, mathematics, computer science, quantitative biology, quantitative finance, statistics, electrical engineering and systems science, and economics.’ =113GB* |
| American Physical Society (APS) | 16B tokens* 1M docs7‘2020 ARTICLES PUBLISHED: 21,845 [Alan: 21,845 x avg 50 years = 1,092,250 docs]’ =47GB* |
| bioRxiv.org – the preprint server for Biology | 4B tokens* 240K docs8‘239,518 Results for posted between “01 Jan, 2010 and 01 Jul, 2024″‘ =11GB* |
| PRO (proprietary dataset mentioned by Argonne, no details) | |
| CORE (proprietary dataset mentioned by Argonne, no details) | |
| ‘Structured narratives’ (generated text that describes data) |
Source: Argonne, 18/Jul/2024
* All token counts are ‘determined,’ based on disclosed data from primary source. Size on disk (uncompressed) uses 1:47 (1M docs = 47GB) as in avg size of arXiv documents @ 1 arXiv doc = 46.61KiB (47.72864 KB). Token count uses 1 arXiv doc = 0.3532 tokens per byte (around 16,857 tokens per doc). Both highlights calculated by EleutherAI for The Pile (31/Dec/2020). Calculations are informed but rough estimates (for example, without internal context, it’s unclear how much of BV-BRC’s 418.3TB/148T tokens of user data is just shared duplication). For similar working see my 2022 paper: What’s in my AI? A Comprehensive Analysis of Datasets Used to Train GPT-1, GPT-2, GPT-3, GPT-NeoX-20B, Megatron-11B, MT-NLG, and Gopher.
There are many well-defined approaches for images. In some cases, there are also approaches for video. However, scientific images and scientific video don’t necessarily look like traditional image sources that are studied widely by the broader community.
For example, if you’re looking at LiDAR data, instead of having three channels like red, green, or blue, it can have up to 128 channels per sensor pixel. If you’re looking at climate simulations, you may have spatial coherence in the X and Y direction, but you may have discontinuities in the Z direction.
These kinds of abnormal aspects of scientific image data present an open challenge that labs like Argonne need to tackle, even though there are established approaches within the scientific community. (Argonne, 18/Jul/2024)
Related datasets: The Well and Multimodal Universe: Comprehensive datasets for scientific research (Dec/2024)
The Well and Multimodal Universe are new extensive datasets designed to support research in machine learning and scientific disciplines. For ML researchers, these two major datasets provide a combined total of 115TB of validated, well-understood scientific data, complete with clean train/test splits and benchmark tasks.
- The Well: 15TB (~4.1T tokens) of physics simulations
The first dataset, “The Well”, contains curated physics simulations from 16 scientific domains, each capturing fundamental equations that appear throughout nature, and all validated and/or generated by domain experts:
- Fluid dynamics & turbulence
- Supernova explosions
- Biological pattern formation
- Acoustic wave propagation
- Magnetohydrodynamics
Available at: https://github.com/PolymathicAI/the_well
- Multimodal Universe: 100TB (~27.5T tokens) of astronomical data
The second dataset, “Multimodal Universe“, contains hundreds of millions of observations across multiple modalities, object types, and wavelengths. The data was collected from JWST, HST, Gaia, and several other major surveys, and unified in a single, ML-ready format.
Available at: https://github.com/MultimodalUniverse/MultimodalUniverse
Compare with other LLM datasets
Open the Datasets Table in a new tab
Video
Preparing Data at Scale for AuroraGPT
Models Table
Summary of current models: View the full data (Google sheets)All dataset reports by LifeArchitect.ai (most recent at top)
| Date | Type | Title |
| Dec/2025 | 📑 | Genesis Mission |
| Jan/2025 | 📑 | What's in Grok? |
| Jan/2025 | 💻 | NVIDIA Cosmos video dataset |
| Aug/2024 | 📑 | What's in GPT-5? |
| Jul/2024 | 💻 | Argonne AuroraGPT |
| Sep/2023 | 📑 | Google DeepMind Gemini: A general specialist |
| Feb/2023 | 💻 | Chinchilla data-optimal scaling laws: In plain English |
| Aug/2022 | 📑 | Google Pathways |
| Mar/2022 | 📑 | What's in my AI? |
| Sep/2021 | 💻 | Megatron the Transformer, and related language models |
| Ongoing... | 💻 | Datasets Table |
Get The Memo
by Dr Alan D. Thompson · Be inside the lightning-fast AI revolution.Informs research at Apple, Google, Microsoft · Bestseller in 147 countries.
Artificial intelligence that matters, as it happens, in plain English.
Get The Memo.
 Alan D. Thompson is a world expert in artificial intelligence, advising everyone from Apple to the US Government on integrated AI. Throughout Mensa International’s history, both Isaac Asimov and Alan held leadership roles, each exploring the frontier between human and artificial minds. His landmark analysis of post-2020 AI—from his widely-cited Models Table to his regular intelligence briefing The Memo—has shaped how governments and Fortune 500s approach artificial intelligence. With popular tools like the Declaration on AI Consciousness, and the ASI checklist, Alan continues to illuminate humanity’s AI evolution. Technical highlights.
Alan D. Thompson is a world expert in artificial intelligence, advising everyone from Apple to the US Government on integrated AI. Throughout Mensa International’s history, both Isaac Asimov and Alan held leadership roles, each exploring the frontier between human and artificial minds. His landmark analysis of post-2020 AI—from his widely-cited Models Table to his regular intelligence briefing The Memo—has shaped how governments and Fortune 500s approach artificial intelligence. With popular tools like the Declaration on AI Consciousness, and the ASI checklist, Alan continues to illuminate humanity’s AI evolution. Technical highlights.This page last updated: 30/Jan/2026. https://lifearchitect.ai/auroragpt/↑
- 1‘Building on a high-performance 1.7PB parallel file system (to be increased in the near future) and embedded in Argonne National Laboratory’s 100+ Gbps network fabric… Given the size of the raw outputs (up to several PBytes for our largest simulations) it is very difficult (and expensive) to provide access to Level 1 data. Level 2 data has an added layer of processing, which makes it roughly an order of magnitude smaller in size; examples include downsampled particle snapshots, halo catalogs, and halo merger trees. Most scientific results are based on the analysis of Level 2 data and post-processing thereof to generate Level 3 data. Level 3 data includes maps and object catalogs at different wavelengths. In this first data release, we focus on Level 2 data, namely downsampled particle outputs and halo catalogs.’
- 2‘Total storage used for user data (TB): 418.3’
- 3‘The piControl run with 500 years of data size: Published 34T of data, 43742 files’
- 4‘PubMed® comprises more than 37 million citations for biomedical literature from MEDLINE, life science journals, and online books. Citations may include links to full text content from PubMed Central and publisher web sites.’
- 5‘As of March 2020, the DL contains 786,000 full-text articles, and 3.2 million tracked publication records.’
- 6‘arXiv is a free distribution service and an open-access archive for nearly 2.4 million scholarly articles in the fields of physics, mathematics, computer science, quantitative biology, quantitative finance, statistics, electrical engineering and systems science, and economics.’
- 7‘2020 ARTICLES PUBLISHED: 21,845 [Alan: 21,845 x avg 50 years = 1,092,250 docs]’
- 8‘239,518 Results for posted between “01 Jan, 2010 and 01 Jul, 2024″‘



